Code
library(tidyverse)
library(tidymodels)
library(factoextra)
data(iris)
iris_data <- iris %>% select(-Species)Using tidymodels
This document demonstrates how to perform clustering in R using the tidymodels framework. Clustering is an unsupervised learning technique that groups similar data points together based on their inherent characteristics. We will use the iris dataset for this demonstration.
First, we load the necessary libraries and the iris dataset.
The Elbow Method is a heuristic used to determine the optimal number of clusters in a dataset. We can visualize the total within-cluster sum of squares as a function of the number of clusters. 3 looks like a good number.
K-Means is a popular clustering algorithm. We will use it to group the iris data into 3 clusters.
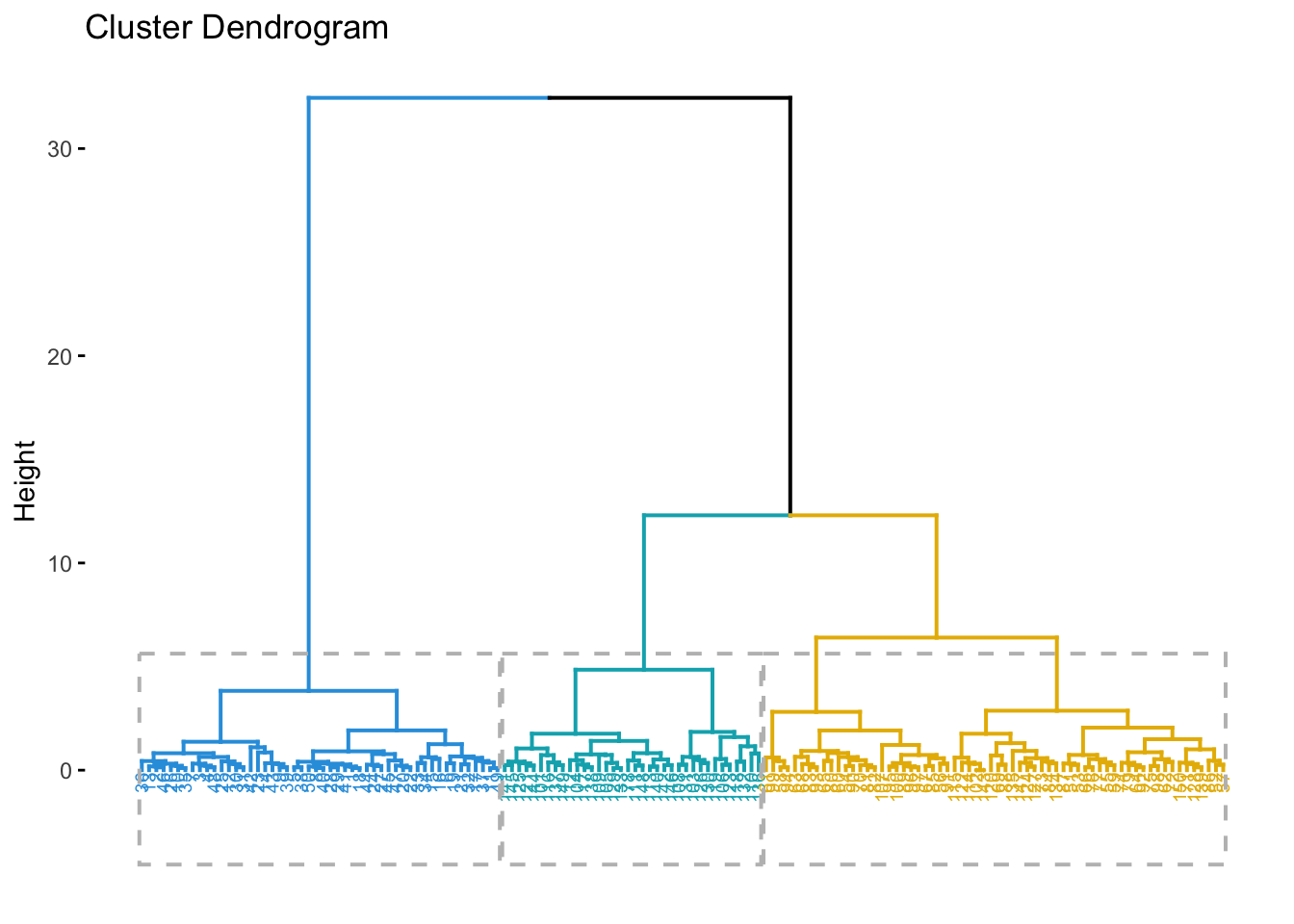
Hierarchical clustering is another common clustering method.
# Calculate the distance matrix
dist_matrix <- dist(iris_data, method = "euclidean")
# Perform hierarchical clustering
hclust_model <- hclust(dist_matrix, method = "ward.D2")
# Visualize the dendrogram
fviz_dend(hclust_model, k = 3, # Cut in 3 groups
cex = 0.5, # label size
k_colors = c("#2E9FDF", "#00AFBB", "#E7B800"),
color_labels_by_k = TRUE, # color labels by groups
rect = TRUE # Add rectangle around groups
)
We can compare the clustering results with the actual species of the iris flowers.
[1] "K-Means Clustering vs. Real Species"
setosa versicolor virginica
1 0 48 14
2 0 2 36
3 50 0 0Here’s a comparison of K-Means and Hierarchical Clustering:
| Feature | K-Means Clustering | Hierarchical Clustering |
|---|---|---|
| Approach | Partitioning (divides data into k clusters) | Agglomerative (bottom-up) or Divisive (top-down) |
| Number of Clusters | Requires pre-specification (k) | Does not require pre-specification; dendrogram helps |
| Computational Cost | Faster for large datasets | Slower for large datasets (O(n^3) or O(n^2)) |
| Cluster Shape | Tends to form spherical clusters | Can discover arbitrarily shaped clusters |
| Sensitivity to Outliers | Sensitive to outliers | Less sensitive to outliers |
| Interpretability | Easy to interpret | Dendrogram can be complex for large datasets |
| Reproducibility | Can vary with initial centroids (unless fixed) | Reproducible |
This document provided a brief overview of clustering in R using tidymodels. We demonstrated both K-Means and Hierarchical clustering on the iris dataset.
---
title: "Clustering:iris data with R"
subtitle: "Using tidymodels"
execute:
warning: false
error: false
format:
html:
toc: true
toc-location: right
code-fold: show
code-tools: true
number-sections: true
code-block-bg: true
code-block-border-left: "#31BAE9"
---
## Introduction
This document demonstrates how to perform clustering in R using the `tidymodels` framework. Clustering is an unsupervised learning technique that groups similar data points together based on their inherent characteristics. We will use the `iris` dataset for this demonstration.
## Load Data
First, we load the necessary libraries and the `iris` dataset.
```{r}
#| label: load-data
#| echo: true
library(tidyverse)
library(tidymodels)
library(factoextra)
data(iris)
iris_data <- iris %>% select(-Species)
```
## Elbow Method
The Elbow Method is a heuristic used to determine the optimal number of clusters in a dataset. We can visualize the total within-cluster sum of squares as a function of the number of clusters. 3 looks like a good number.
```{r}
#| label: elbow-method
#| echo: true
fviz_nbclust(iris_data, kmeans, method = "wss") +
labs(subtitle = "Elbow Method")
```
## K-Means Clustering
K-Means is a popular clustering algorithm. We will use it to group the iris data into 3 clusters.
```{r}
#| label: kmeans
#| echo: true
set.seed(123)
kmeans_model <- kmeans(iris_data, centers = 3, nstart = 25)
# Visualize the clusters
fviz_cluster(kmeans_model, data = iris_data)
```
## Hierarchical Clustering
Hierarchical clustering is another common clustering method.
```{r}
#| label: hclust
#| echo: true
# Calculate the distance matrix
dist_matrix <- dist(iris_data, method = "euclidean")
# Perform hierarchical clustering
hclust_model <- hclust(dist_matrix, method = "ward.D2")
# Visualize the dendrogram
fviz_dend(hclust_model, k = 3, # Cut in 3 groups
cex = 0.5, # label size
k_colors = c("#2E9FDF", "#00AFBB", "#E7B800"),
color_labels_by_k = TRUE, # color labels by groups
rect = TRUE # Add rectangle around groups
)
```
## Comparison with Real Groups
We can compare the clustering results with the actual species of the iris flowers.
```{r}
#| label: comparison
#| echo: true
# K-Means Comparison
iris$kmeans_cluster <- kmeans_model$cluster
print("K-Means Clustering vs. Real Species")
table(iris$kmeans_cluster, iris$Species)
```
```{r}
# Hierarchical Clustering Comparison
iris$hclust_cluster <- cutree(hclust_model, k = 3)
print("Hierarchical Clustering vs. Real Species")
table(iris$hclust_cluster, iris$Species)
```
## Comparison of K-Means and Hierarchical Clustering
Here's a comparison of K-Means and Hierarchical Clustering:
| Feature | K-Means Clustering | Hierarchical Clustering |
|:--------------------|:-------------------------------------------------|:------------------------------------------------------|
| **Approach** | Partitioning (divides data into k clusters) | Agglomerative (bottom-up) or Divisive (top-down) |
| **Number of Clusters** | Requires pre-specification (k) | Does not require pre-specification; dendrogram helps |
| **Computational Cost** | Faster for large datasets | Slower for large datasets (O(n^3) or O(n^2)) |
| **Cluster Shape** | Tends to form spherical clusters | Can discover arbitrarily shaped clusters |
| **Sensitivity to Outliers** | Sensitive to outliers | Less sensitive to outliers |
| **Interpretability** | Easy to interpret | Dendrogram can be complex for large datasets |
| **Reproducibility** | Can vary with initial centroids (unless fixed) | Reproducible |
## Conclusion
This document provided a brief overview of clustering in R using `tidymodels`. We demonstrated both K-Means and Hierarchical clustering on the `iris` dataset.